Quickstart + Usage¶
After installing Consenrich, you can run it via the command line (consenrich -h) or programmatically using the Python/Cython API.
We provide several usage examples below. See also File Formats and Miscellaneous Guidance for more information.
Getting Started: Minimal Example¶
A brief analysis using H3K27ac (narrow mark) ChIP-seq data is carried out for demonstration.
Input Data¶
The input data in this example consists of four donors’ treatment and control samples (epidermal tissue) from ENCODE.
Experiment |
Biosample |
H3K27ac Alignment |
Control Alignment |
|---|---|---|---|
Epidermis/Female/71 |
|||
Epidermis/Male/67 |
|||
Epidermis/Female/80 |
|||
Epidermis/Male/75 |
Download Alignment Files from ENCODE¶
Copy+paste the following to your terminal to download and index the BAM files for this demo.
You can also use curl -O <URL> in place of wget <URL> if the latter is not available on your system.
encodeFiles=https://www.encodeproject.org/files
for file in ENCFF793ZHL ENCFF647VPO ENCFF809VKT ENCFF295EFL; do
wget "$encodeFiles/$file/@@download/$file.bam"
done
for ctrl in ENCFF444WVG ENCFF619NYP ENCFF898LKJ ENCFF490MWV; do
wget "$encodeFiles/$ctrl/@@download/$ctrl.bam"
done
samtools index -M *.bam
Using a YAML Configuration file¶
Tip
Refer to the <process,observation,etc.>Params classes in module in the API for complete documentation of configuration options.
Copy and paste the following YAML into a file named demoHistoneChIPSeq.yaml. For a quick trial run (\(\approx\) 1 minute), you can restrict analysis to a subset of chromosomes: To reproduce the results shown in the browser snapshot, add genomeParams.chromosomes: [chr21, chr22] to the configuration file.
# v0.7.11b1
experimentName: demoHistoneChIPSeq
genomeParams.name: hg38
genomeParams.chromosomes: [chr21, chr22] # remove this line to run genome-wide
genomeParams.excludeForNorm: [chrX, chrY]
inputParams.bamFiles: [ENCFF793ZHL.bam,
ENCFF647VPO.bam,
ENCFF809VKT.bam,
ENCFF295EFL.bam]
inputParams.bamFilesControl: [ENCFF444WVG.bam,
ENCFF619NYP.bam,
ENCFF898LKJ.bam,
ENCFF490MWV.bam]
# Optional: call 'structured peaks'
matchingParams.templateNames: [haar, db2]
matchingParams.cascadeLevels: [3,3]
matchingParams.methodFDR: BH
outputParams.writeStateStd: true
Control Inputs
Omit inputParams.bamFilesControl for ATAC-seq, DNase-seq, Cut&Run, and other assays where no control is available or applicable.
Run Consenrich¶
Guidance: Command-line vs. Programmatic Usage
The command-line interface is a convenience wrapper that may not expose all available objects or more niche features. Some users may find it beneficial to run Consenrich programmatically (e.g., in a Jupyter notebook, Python script), as the API enables greater flexibility to apply custom preprocessing steps and various context-specific protocols within existing workflows.
% consenrich --config demoHistoneChIPSeq.yaml --verbose
Results¶
We display Consenrich results (blue) at
APOL2 <--| |--> APOL1For reference, ENCODE peaks (label: rep1 pseudoreplicated peaks) for the same Experiments and donor samples are included (black):
ENCODE Histone ChIP-seq pipeline (unreplicated) (MACS peak calls, partition concordance)

Additional Examples and Benchmarking¶
This section of the documentation will be regularly updated to include a breadth of assays, downstream analyses, and runtime benchmarks.
ATAC-seq¶
Input data (atac20): \(m=20\) ATAC-seq BAM files derived from lymphoblastoid cell lines (ENCODE)
Varying data quality (e.g., Extremely low read depth)
Environment¶
MacBook MX313LL/A (arm64)
Python 3.12.9
Consenrich v0.7.9b1
HTSlib (Samtools) 1.22.1
Bedtools 2.31.1
Names and versions of packages that are relevant to computational performance. These specific versions are not required but are included for reproducibility.
Package |
Version |
|---|---|
|
3.1.4 |
|
2.3.3 |
|
1.16.2 |
Configuration¶
Run with the following YAML config file atac20Benchmark.yaml. Note that several parameters are listed and/or adjusted for demonstration purposes.
globs, e.g., *.bam, are allowed, but each file is listed below to reveal ENCODE accessions
experimentName: atac20Benchmark
genomeParams.name: hg38
genomeParams.excludeChroms: ['chrY']
genomeParams.excludeForNorm: ['chrX', 'chrY']
inputParams.bamFiles: [
ENCFF326QXM.bam,
ENCFF497QOS.bam,
ENCFF919PWF.bam,
ENCFF447ZRG.bam,
ENCFF632MBC.bam,
ENCFF949CVL.bam,
ENCFF462RHM.bam,
ENCFF687QML.bam,
ENCFF495DQP.bam,
ENCFF767FGV.bam,
ENCFF009NCL.bam,
ENCFF110EWQ.bam,
ENCFF797EAL.bam,
ENCFF801THG.bam,
ENCFF216MFD.bam,
ENCFF588QWF.bam,
ENCFF795UPB.bam,
ENCFF395ZMS.bam,
ENCFF130DND.bam,
ENCFF948HNW.bam
]
# Optional: call 'structured peaks' via `consenrich.matching`
matchingParams.templateNames: [haar, db2]
matchingParams.cascadeLevels: [3, 3]
matchingParams.minMatchLengthBP: -1
matchingParams.mergeGapBP: -1
# Optional: plot distributions
plotParams.plotStateTrace: true
plotParams.plotResidualsHistogram: true
plotParams.plotStateStdHistogram: true
countingParams.applySqrt: true
Run Consenrich¶
% consenrich --config atac20Benchmark.yaml --verbose
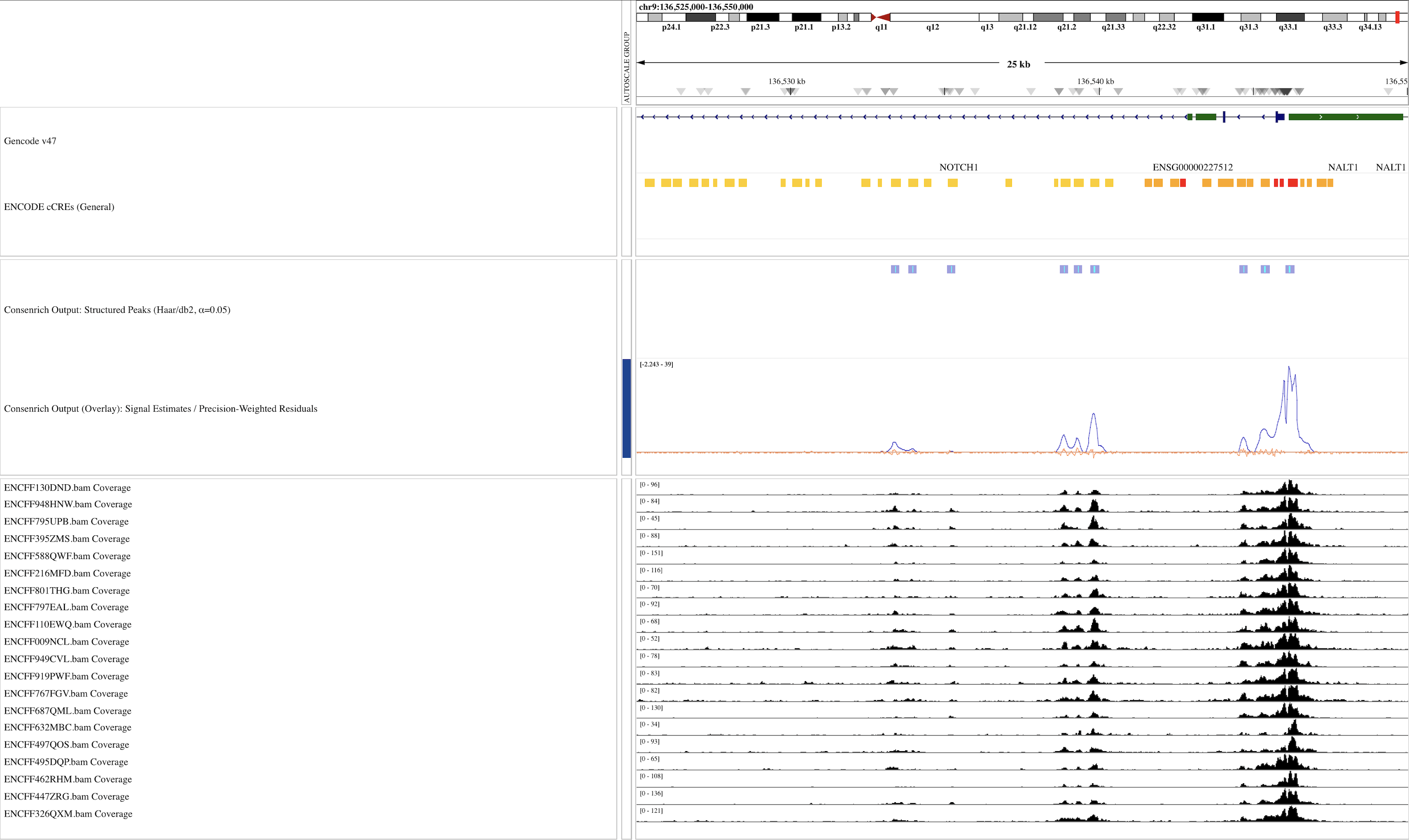
Results¶
Browser Snapshot
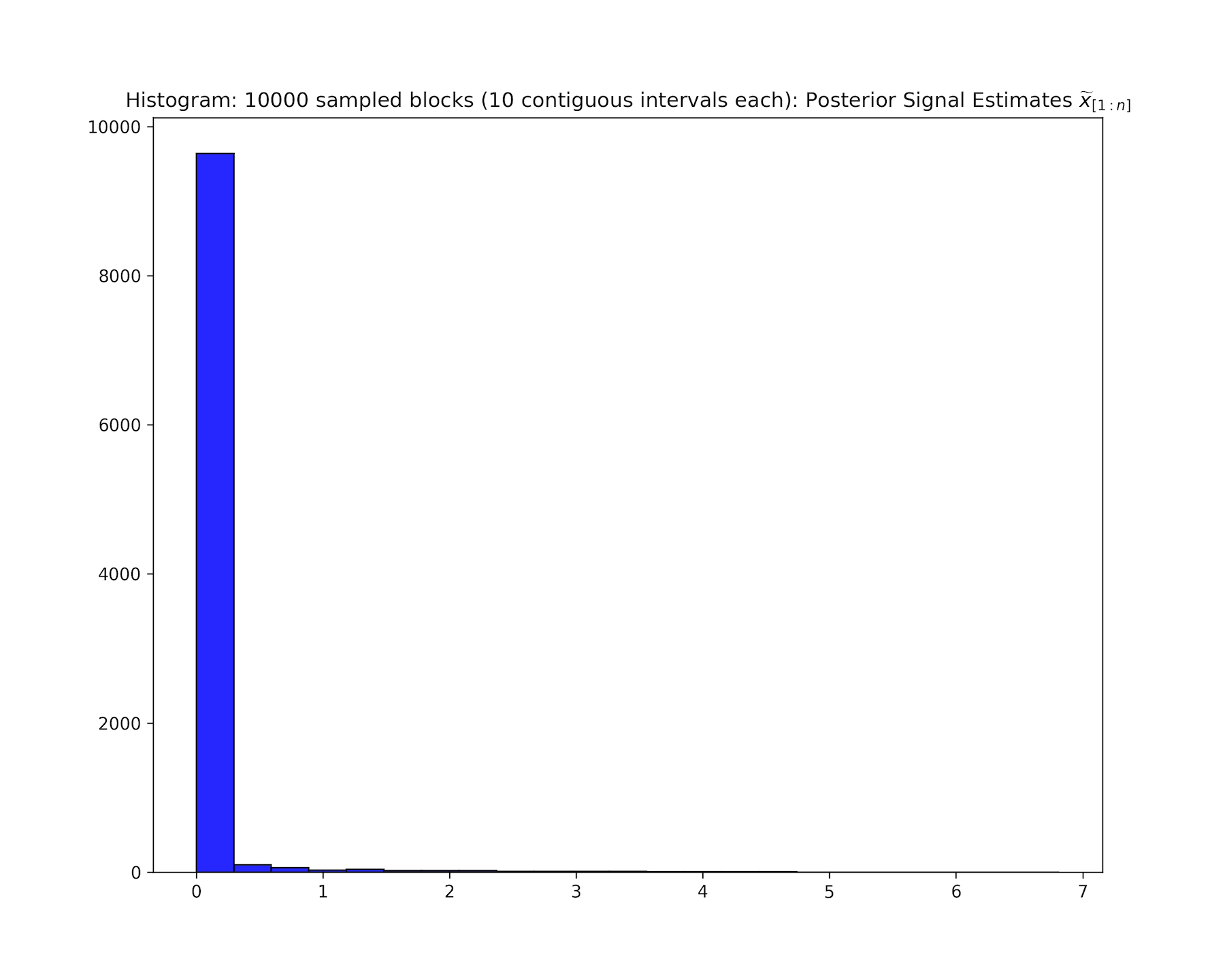
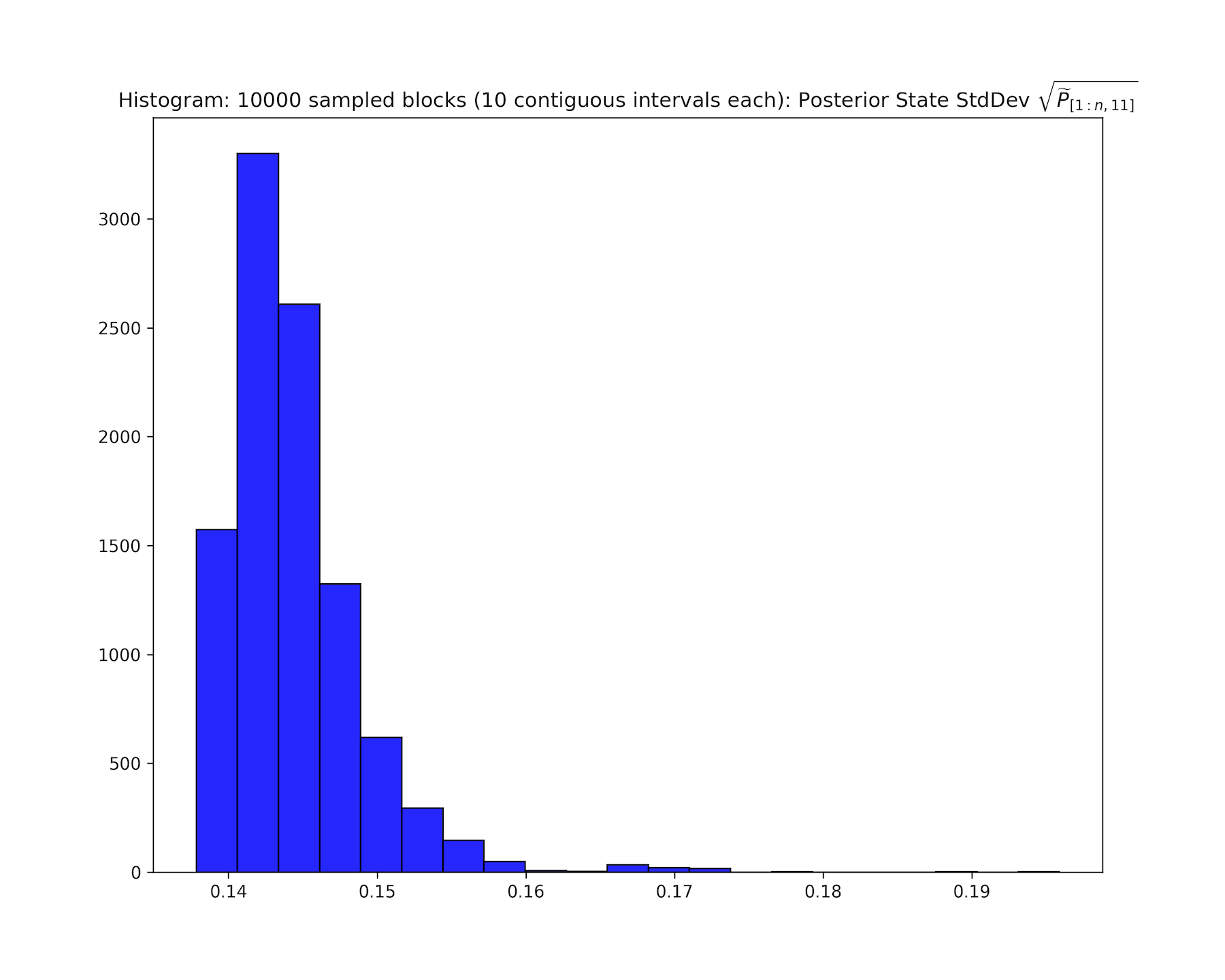
Visualizing State and Residual Distributions
For downstream analyses, it can be useful to inspect the distributions of state estimates, residuals, and state uncertainties.
Basic convenience utilities are provided for this purpose. Here, we display plots corresponding to chr19, as in the browser snapshot above. Note, plotParams.plot<...> will create a separate directory <experimentName>_consenrichPlots to store files.
See consenrich.core.plotParams and its associated functions for more details.
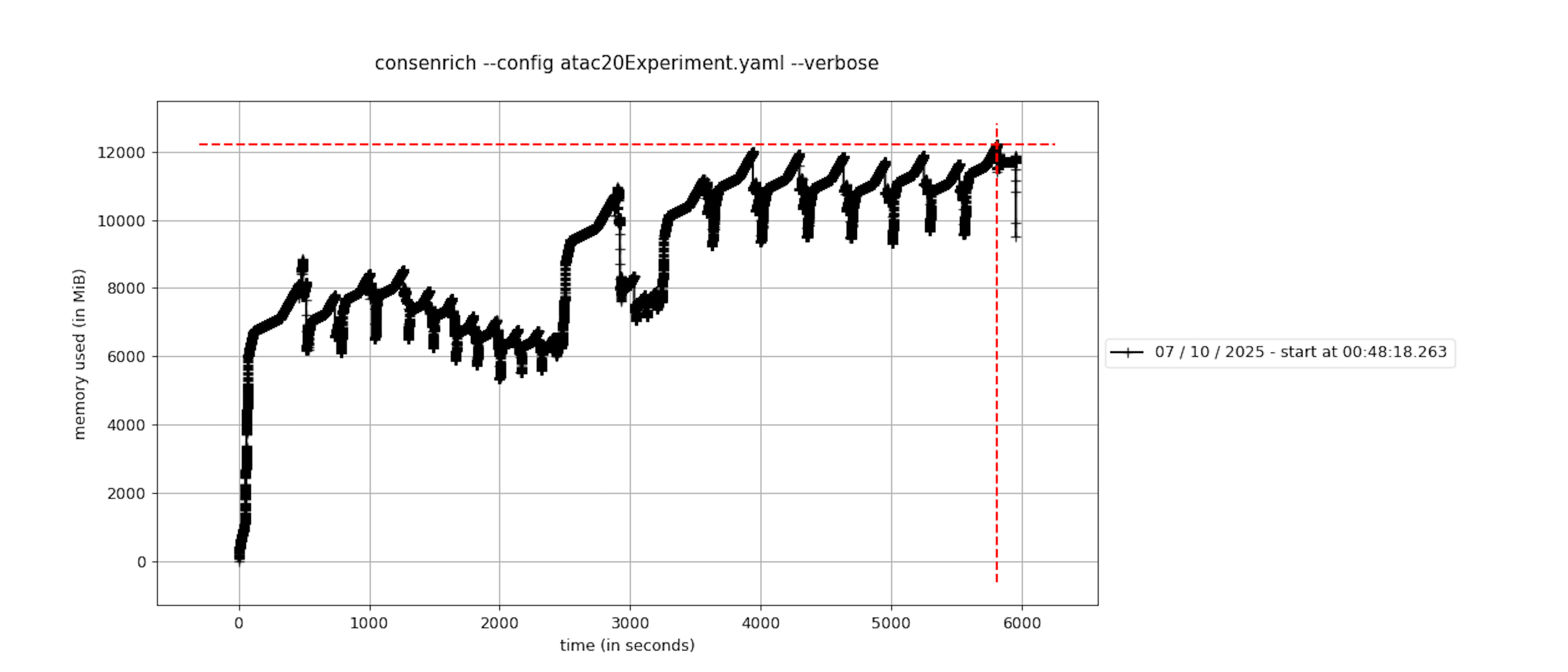
Runtime and Memory Profiling¶
Memory was profiled using the package memory-profiler. See the plot below for memory usage over time. Function calls are marked as notches.
Note that the repeated sampling of memory every 0.1 seconds during profiling introduces some overhead that affects runtime. MacOS’s caffeinate command was used to prevent sleeping during execution.
ChIP-seq: Broad Histone Marks¶
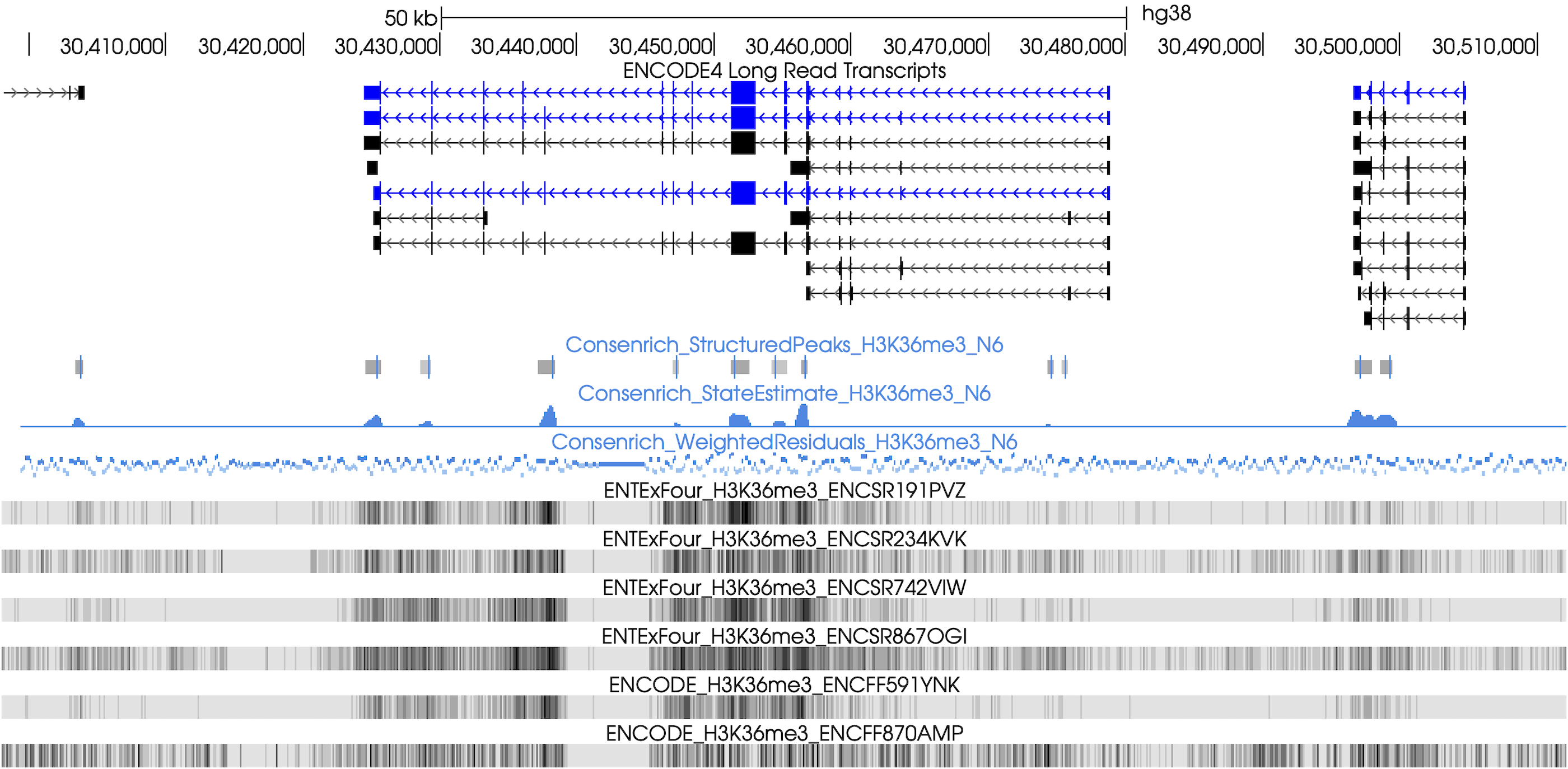
In this demo, we use \(m=6\) H3K36me3 ChIP-seq samples from separate donors’ lung tissues.
Twelve total alignment files (single-end, treatment/control input pairs) are used. See the YAML below for ENCFF<fileID> accessions.
Configuration¶
experimentH3K36me3.yaml.experimentName: experimentH3K36me3 genomeParams.name: hg38 genomeParams.excludeChroms: [chrY] genomeParams.excludeForNorm: [chrX, chrY] inputParams.bamFiles: [ENCFF441SHP.bam, ENCFF450ORQ.bam, ENCFF903UTS.bam, ENCFF790HIV.bam, ENCFF591YNK.bam, ENCFF870AMP.bam ] inputParams.bamFilesControl: [ENCFF794QJK.bam, ENCFF831MFQ.bam, ENCFF660HBS.bam, ENCFF430OFG.bam, ENCFF347ENG.bam, ENCFF648HNK.bam ] # include longer more symmetric `sym3` template for broad marks matchingParams.templateNames: [haar, sym3] matchingParams.cascadeLevels: [3, 3] matchingParams.minMatchLengthBP: -1 matchingParams.mergeGapBP: 500 # increase merge radius for broad marks countingParams.applySqrt: true
Run Consenrich¶
% consenrich --config experimentH3K36me3.yaml --verbose
Results¶
Signal estimates, weighted residuals, and structured peaks (via `matching`) over a large genomic region spanning LINC01176, NOD1, GGCT:
File Formats¶
Input
Per-sample sequence alignment files (BAM format)
Optional: Control/input alignment files (e.g., ChIP-seq)
Note, if using Consenrich programmatically, users can provide preprocessed sample-by-interval count matrices directly instead of BAM files (see
consenrich.core.runConsenrich())
Output
Posterior Signal estimate track:
<experimentName>_consenrich_state.bwThis track records genome-wide Consenrich estimates for the targeted signal of interest
A human-readable bedGraph file is also generated:
consenrichOutput_<experimentName>_consenrich_state.bedGraph
Weighted post-fit residual track:
<experimentName>_consenrich_residuals.bwThis track records genome-wide differences between (a) Consenrich estimates and (b) observed sample data – after accounting for regional + sample-specific uncertainty.
These values can reflect model mismatch: Where they are large (magnitude), the model’s estimated uncertainty may fail to explain discrepancies with the observed data.
A human-readable bedGraph file is also generated:
consenrichOutput_<experimentName>_consenrich_residuals.bedGraph
Posterior state uncertainty track:
<experimentName>_consenrich_stateStd.bwPointwise uncertainty in the primary state estimate, \(\sqrt{\widetilde{P}_{i,(11)}}\), on a scale comparable to the estimated signal.
A human-readable bedGraph file is also generated:
consenrichOutput_<experimentName>_consenrich_stateStd.bedGraph
Structured peak calls (Optional):
<experimentName>_matches.mergedMatches.narrowPeakBED-like annotation of enriched signal regions showing a regular structure. Only generated if the matching algorithm is invoked.
See outputParams in the API for full documentation of output options.
Miscellaneous Guidance¶
Consensus Peak Calling + Downstream Differential Analyses¶
Consenrich can improve between-group differential analyses that depend on a good set of initial ‘candidate’ consensus peaks (see Enhanced Consensus Peak Calling and Differential Analyses in Complex Human Disease in the manuscript preprint.)
ROCCO can accept Consenrich bigWig files as input and is well-suited to leverage high-resolution open chromatin signal estimates while balancing regularity for simultaneous broad/narrow peak calling.
For example, to run the Consenrich+ROCCO protocol as it is used in the manuscript,
% python -m pip install rocco --upgrade
% rocco -i <experimentName>_consenrich_state.bw \
-g hg38 -o consenrichRocco_<experimentName>.bed \
# <...>
The budgeted/total-variation-regularized optimization procedure performed by ROCCO to select consensus peak regions prevents excessive multiple comparisons downstream and enforces biological plausibility. Other peak calling methods—including the `matching` algorithm packaged with Consenrich—that accept bedGraph or bigWig input (e.g., MACS’ bdgpeakcall) may also prove viable, but only Consenrich+ROCCO has been extensively benchmarked for differential accessibility analyses to date.
In general, for workflows of the form Consenrich Signal Track --> Peak Caller --> Sample-by-CalledPeaks Count Matrix --> Differential Analysis between Conditions, it is recommended to use all samples from all experimental conditions as input to Consenrich for better control of downstream false discovery rates. See, for example, Lun and Smyth, 2014.
Matching Algorithm: Command-line Usage¶
To avoid a full run/rerun of Consenrich when calling structured peaks, the matching algorithm can be run directly at the command-line on existing Consenrich-generated bedGraph files. For example:
% consenrich \
--match-bedGraph consenrichOutput_<experimentName>_state.bedGraph \
--match-template sym3 sym4 \
--match-level 2 2 \
--match-alpha 0.01
Run ``consenrich -h`` for additional command-line options.
For more details on the matching algorithm in general, see `matching` and consenrich.matching.matchWavelet().
Broad, Heterochromatic and/or Repressive targets¶
Several options are available for targeting broad, domain-level features:
Decrease noise-vulnerability with larger genomic intervals (
countingParams.stepSize). The default is 25bp, but increasing up to 250 bp or more may help stabilize estimates for very broad domainsSoften or disable background-removal/detrending (
detrendParams) to avoid removing important broad/low-frequency signalInclude larger/more symmetric wavelet/scaling templates (e.g., sym3, sym4) and larger
matchingParams.mergeGapBPfor the matching algorithm (`matching`)
If the targeted mark is associated with silencing/repression/heterochromatin, the default genomeParams.sparseBedFile annotations packaged with Consenrich may not be appropriate. Consider invoking the annotation-free variant of the ALV method (consenrich.core.getAverageLocalVariance()) with observationParams.useALV: true or specifying a custom genomeParams.sparseBedFile
Preprocessing and Calibration of Uncertainty Metrics¶
To promote homoskedasticity, symmetry, independence, etc. of residuals for downstream analyses that require calibrated uncertainty quantification it is often helpful/necessary to transform count-based data.
Consenrich’s uncertainty estimates can generally be interpreted on a relative scale (e.g. comparing between genomic regions), but classic statistical interpretations, e.g., coverage of prediction intervals, can be made more reliable through appropriate preprocessing.
Log-like transforms (
applyAsinh:=numpy.arcsinh,applyLog:=numpy.log1p) are useful for stripping multiplicative noise components for additive linear modeling. Depending on sparsity, their comparably strong compression may affect capture of subtle signal patterns when applying Consenrich.applySqrtoffers a comparatively gentle compression of the dynamic range and may be preferable to log transforms if needing to preserve a greater breadth of signal variation for downstream tasks like peak calling. This transformation may not be fully variance-stabilizing for highly overdispersed data, however.Users running Consenrich programmatically can apply custom transformations and preprocesssing pipelines as desired, e.g., Yeo-Johnson or general power transforms.
Note, in the default (CLI) implementation, these transformations are applied before detrending (consenrich.detrorm).