ROCCO: [R]obust [O]pen [C]hromatin Detection via [C]onvex [O]ptimization¶
What¶
ROCCO is an efficient algorithm for detection of “consensus peaks” in large datasets with multiple HTS data samples (namely, ATAC-seq), where an enrichment in read counts/densities is observed in a nontrivial subset of samples.
Input/Output¶
Input: Samples’ BAM alignments or BigWig tracks
Output: BED (or narrowPeak) file of consensus peak regions
Note, if BigWig input is used, no preprocessing options can be applied at the alignment level.
How¶
ROCCO models consensus peak calling as a constrained optimization problem with an upper-bound on the total proportion of the genome selected as open/accessible and a fragmentation penalty to promote spatial consistency in active regions and sparsity elsewhere.
Why¶
ROCCO offers several attractive features:
Consideration of enrichment and spatial characteristics of open chromatin signals
Scaling to large sample sizes (100+) with an asymptotic time complexity independent of sample size
Unsupervised Does not require training data or a heuristically determined set of initial candidate peak regions
No rigid thresholds on the minimum number/width of supporting samples/replicates
Mathematically tractable model permitting worst-case analysis of runtime and performance
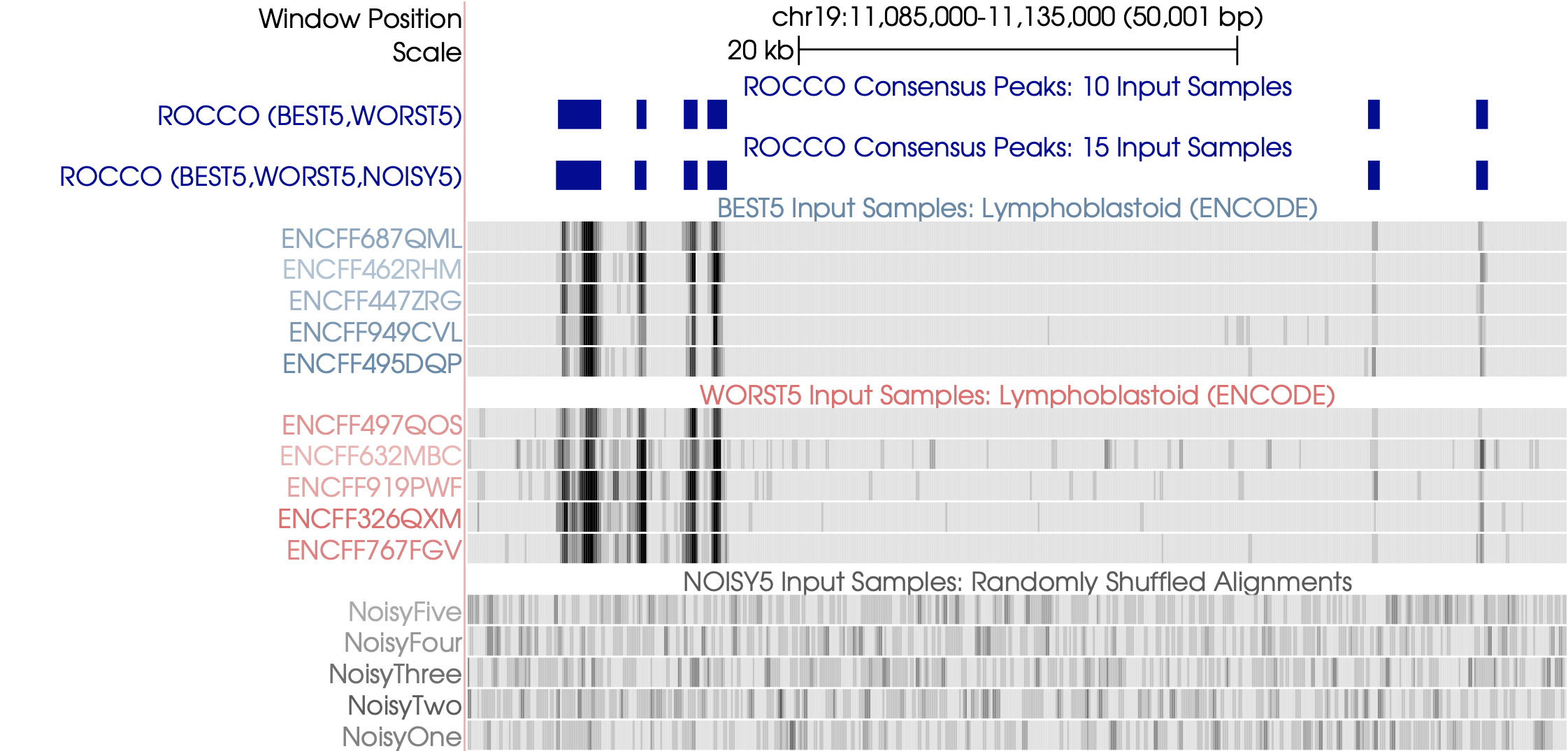
Example Behavior¶
Input¶
ENCODE lymphoblastoid data (BEST5, WORST5): 10 real ATAC-seq alignments of varying TSS enrichment (SNR-like)
Synthetic noisy data (NOISY5)
We run twice under two conditions – once with noisy samples and once without to compare results (blue).
Ideally, the resulting consensus peaks should be largely unaffected by the Noisy5 input samples. Likewise, both broad and narrow peaks should be captured.
rocco -i *.BEST5.bam *.WORST5.bam -g hg38 -o rocco_output_without_noise.bed
rocco -i *.BEST5.bam *.WORST5.bam *.NOISY5.bam -g hg38 -o rocco_output_with_noise.bed
Note, users may run ROCCO with flag –narrowPeak to generate 10-column output with various statistics for comparison.
Output¶
Paper/Citation¶
If using ROCCO in your research, please cite the original paper in Bioinformatics (DOI: btad725)
Nolan H Hamilton, Terrence S Furey, ROCCO: a robust method for detection of open chromatin via convex optimization,
Bioinformatics, Volume 39, Issue 12, December 2023
Documentation¶
For additional details, usage examples, etc. please see ROCCO’s documentation: https://nolan-h-hamilton.github.io/ROCCO/
Installation¶
PyPI (pip)¶
python -m pip install rocco --upgrade
If lacking administrative control, you may need to append –user to the above.
Build from Source¶
If preferred, ROCCO can easily be built from source:
Clone or download this repository
git clone https://github.com/nolan-h-hamilton/ROCCO.git cd ROCCO python setup.py sdist bdist_wheel pip install -e .
System-Level Dependencies:¶
ROCCO utilizes the popular bioinformatics software Samtools (http://www.htslib.org) and bedtools (https://bedtools.readthedocs.io/en/latest/).
If these are not already available, you can install system-wide dependencies with a package manager e.g.,
For Homebrew (MacOS):
brew install samtools
brew install bedtools
For Ubuntu/Debian:
sudo apt-get install samtools
sudo apt-get install bedtools
Conda:
conda install bioconda::bedtools
conda install bioconda::samtools
or built from source (See Samtools (http://www.htslib.org) and bedtools (https://bedtools.readthedocs.io/en/latest/)).
As a supplementary resource, see docs/environments in the GitHub repository for environment files with detailed package version information.
- chrom_solution_to_bed(chromosome, intervals, solution, ID=None, check_gaps_intervals=True, min_length_bp=None) str[source]¶
Convert the ROCCO-generated vector of decision variables for a given chromosome to a BED file
- Parameters:
chromosome (str) – Chromosome name
intervals (np.ndarray) – Intervals for the chromosome
solution (np.ndarray) – Solution vector for the chromosome
ID (str) – Unique identifier for the solution
check_gaps_intervals (bool) – Check if intervals are contiguous and fixed width
min_length_bp (int) – Minimum length of a region to be included in the output BED file
- Returns:
Output BED file
- Return type:
str
- combine_chrom_results(chrom_bed_files: list, output_file: str, name_features: bool = False) str[source]¶
Combine the results from individual chromosome solutions into a single BED file after running ROCCO on each chromosome
- Parameters:
chrom_bed_files (list) – List of BED files for each chromosome
output_file (str) – Output BED file
name_features (bool) – Name the features in the output BED file
- Returns:
Output BED file
- Return type:
str
- cscores_quantiles(chrom_scores: numpy.ndarray, quantiles: numpy.ndarray | None = None, add_newlines=True) str[source]¶
Returns a formatted string of quantiles for a given array of ‘locus scores’ (
score_chrom_linear())- Parameters:
chrom_scores (np.ndarray) – locus scores (float) for a given chromosome
quantiles (np.ndarray, optional) – array of quantiles in [0.0,1.0] to compute.
- Returns:
pformatted string of quantiles
- Return type:
str
- Seealso:
- get_floor_eps_sol(chrom_lp_sol: numpy.ndarray, budget: float, int_tol: float = 1e-06, eps_mult: float = 1.01) numpy.ndarray[source]¶
Compute the integral floor_eps solution from the relaxation
Adds a small \(\epsilon\) to each decision variable before applying the floor function. Addresses solutions in the interior of the feasible region near integer corner point solutions. The floor_eps approach is a crude heuristic to obtain an initial integer-feasible solution from the LP relaxation, but is well-suited as an initial reference point for randomization (ROCCO-RR). In many cases, the relaxed LP solution is nearly integral and the floor_eps heuristic and randomized ROCCO-RR procedure has little effect.
- Parameters:
chrom_lp_sol (np.ndarray) – Solution vector from the LP relaxation
budget (float) – \(b\) upper bounds the proportion of the chromosome that can be selected as open/accessible
int_tol (float) – If a decision variable is within int_tol of 0 or 1, it is considered integral
eps_mult (float) – Value by which to divide the initial epsilon value
- Returns:
Initial integer-feasible solution
- Return type:
np.ndarray
- get_rround_sol(chrom_lp_sol, scores, budget, gamma, rand_iter=1000, int_tol=1e-06, eps_mult: float = 1.01) Tuple[numpy.ndarray, float][source]¶
Get the ROCCO-RR solution from the solution to the relaxation
- Parameters:
chrom_lp_sol (np.ndarray) – Solution vector from the LP relaxation
scores (np.ndarray) – Scores for each genomic position within a given chromosome
budget (float) – \(b\) upper bounds the proportion of the chromosome that can be selected as open/accessible
gamma (float) – \(\gamma\) is the coefficient for the fragmentation penalty used to promote spatial consistency in distinct open genomic regions and sparsity elsewhere.
rand_iter (int) – Number of randomizations to obtain the ROCCO-RR solution
int_tol (float) – If a decision variable is within int_tol of 0 or 1, it is considered integral and ignored in the randomization step
eps_mult (float) – Value by which to divide the initial
get_floor_eps_sol()epsilon value
- Returns:
ROCCO-RR solution and optimal value
- Return type:
Tuple[numpy.ndarray, float]
- parsig(scores, gamma=None, parsig_B=None, parsig_M=None, parsig_R=None, scale_quantile: float = 0.5, remove_min=True) numpy.ndarray[source]¶
Applies a ‘parametric sigmoid’ (smooth step) function mapping input scores to [0,M)
- Parameters:
parsig_M (float) – Upper bound (sup.) of the transformed score. If None, determined by scale_quantile and gamma
parsig_B (float) – Determines the inflection past which values are mapped closer to M. An inflection point occurs in the transformation at quantile(scores, parsig_B)/2. For a given parsig_R, larger parsig_B value will result in more scores being pushed toward the minimum value of the transformed scores.
parsig_R (float) – Scales the rate of change around the step.
scale_quantile (float)
- Return type:
np.ndarray
- resolve_config(args)[source]¶
Resolve command-line arguments with a JSON configuration file
- Parameters:
args (dict) – Command-line arguments obtained with argparse
- Returns:
Resolved command-line arguments
- Return type:
dict
Note
Modifies/overrides command-line arguments specified explicitly in the JSON file
For boolean arguments, use true or false in the JSON file rather than True or False
Example JSON config file
{ "input_files": ["sample1.bam", "sample2.bam"], "output": "rocco_peaks_output.bed", "genome": "hg38", "chroms": ["chr21", "chr22"], "int_tol": 0.01, "verbose": true, "solver": "pdlp" }
Can then run rocco –config config.json […].
- resolve_transformations(args: dict)[source]¶
Check if both savgol and medfilt transformations are invoked by CLI, resolve to medfilt if so.
- Parameters:
args (dict) – Command-line arguments
- Returns:
Resolved command-line arguments
- Return type:
dict
- score_boundary_chrom(signal_vector: numpy.ndarray, denom: float = 1.0, power: float = 1.0, smooth_window: int = -1, smooth_degree: int = 3) numpy.ndarray[source]¶
Calculate the boundary stats for the chromosome score (maximum absolute difference in either direction)
- Parameters:
signal_vector (np.ndarray) – (Assumed: central tendency stats) vector for a given chromosome
denom (float) – Denominator for boundary stats
power (float) – Power to raise the boundary stats to
smooth_window (int) – If > 1, smooth as a polynomial function fit to local windows of this length (Savitzky-golay filter)
smooth_degree (int) – Polynomial degree for SG filter.
- Returns:
Boundary scores
- Return type:
np.ndarray
- score_central_tendency_chrom(chrom_matrix, method='quantile', quantile=0.5, tprop=0.05, power=1.0) numpy.ndarray[source]¶
Calculate the central tendency of read measures across samples (rows) at each genomic position (column) in a given chrom.
- Parameters:
chrom_matrix (np.ndarray) – Matrix of values for a given chromosome
method (str) – Method to calculate central tendency. Options are ‘quantile’, ‘tmean’ (trimmed-mean), and ‘mean’. Default is quantile, q=.50 (median)
quantile (float) – Quantile to use if method is ‘quantile’
tprop (float) – Proportion of values to trim if method is ‘tmean’
power (float) – Power to raise the central tendency value to
- Returns:
Central tendency score
- Return type:
np.ndarray
- score_chrom_linear(central_tendency_vec: numpy.ndarray, dispersion_vec: numpy.ndarray, boundary_vec: numpy.ndarray, gamma=None, c_1=1.0, c_2=-1.0, c_3=1.0, eps_neg=-0.001, transform_parsig=False, parsig_B=None, parsig_M=None, parsig_R=None) numpy.ndarray[source]¶
Compute array of default scores \((\mathbf{G}\boldsymbol{\alpha})^{\top}\) where \(\mathbf{G}\) is the \(n \times 3\) matrix of central tendency scores, dispersion scores, and boundary scores for a given chromosome and \(\boldsymbol{\alpha}\) is the 3D vector of coefficients for each.
- Parameters:
central_tendency_vec (np.ndarray) – Central tendency scores for a given chromosome
dispersion_vec (np.ndarray) – Dispersion scores for a given chromosome
boundary_vec (np.ndarray) – Boundary scores for a given chromosome
gamma (float) – \(\gamma\) is the coefficient for the fragmentation penalty used to promote spatial consistency in distinct open genomic regions and sparsity elsewhere. The ideal value of gamma is for a given dataset is dependent on the user’s preference. Increase to encourage solutions that are ‘block-like’.
c_1 (float) – Coefficient for central tendency scores (\(c_1 g_1(i),~i=1,\ldots,n\) in the paper)
c_2 (float) – Coefficient for dispersion scores (\(c_2 g_2(i),~i=1,\ldots,n\) in the paper)
c_3 (float) – Coefficient for boundary scores (\(c_3 g_3(i),~i=1,\ldots,n\) in the paper)
parsig_M (float) – Upper bound (sup.) of scores under the transformation
parsig_B (float) – An inflection point occurs in the transformation at quantile(scores, parsig_B)/2. For a given parsig_R, larger parsig_B value will result in more scores being pushed toward the minimum value of the transformed scores.
parsig_R (float) – Scales the rate of change around the inflection point at quantile(scores, parsig_B)/2. Higher values of parsig_R will result in a steeper sigmoid curve, approaching a step function in the limit with two distinct values.
eps_neg (float) – Negative epsilon value to add to the scores
- Returns:
chromosome scores
- Return type:
np.ndarray
- score_dispersion_chrom(chrom_matrix: numpy.ndarray, method: str = 'mad', rng=(25, 75), tprop=0.05, power: float = 1.0) numpy.ndarray[source]¶
Calculate the dispersion of read measures across samples (rows) at each genomic position (column) in a given chrom.
- Parameters:
chrom_matrix (np.ndarray) – Matrix of values for a given chromosome
method (str) – Method to calculate dispersion. Options are ‘mad’, ‘iqr’, ‘tstd’ (trimmed std.), and ‘std’. Default is ‘mad’
tprop (float) – Proportion of values to trim if method is ‘tstd’
rng (Tuple[int,int]) – Range of quantiles to use if method is ‘iqr’
power (float) – Power to raise the dispersion value to
- Returns:
Dispersion scores
- Return type:
np.ndarray
- solve_relaxation_chrom_glop(scores, budget: float = 0.035, gamma: float = 1.0, beta: float | None = None, denom_const: float | None = None, glop_parameters=None, glop_dual_feasibility_tolerance: float = 1e-06, glop_primal_feasibility_tolerance: float = 1e-06, glop_use_scaling: bool = True, glop_initial_basis: str = 'TRIANGULAR', glop_push_to_vertex: bool = True, glop_use_dual_simplex=None, glop_allow_simplex_algorithm_change: bool = True, scale_gamma: bool = False, hint=None, threads: int = 0, verbose: bool = False, save_model: str | None = None) Tuple[numpy.ndarray, float][source]¶
Solve the relaxation for a specific chromosome using the simplex-based method, glop
OR-tools linear programming resources and documentation
A simplex-based method that yields corner-point feasible (CPF, vertex) solutions. May become deprecated in future releases of ROCCO in favor of the pdlp solver.
- Parameters:
scores (np.ndarray) – Scores for each genomic position within a given chromosome
budget (float) – \(b\) upper bounds the proportion of the chromosome that can be selected as open/accessible
gamma (float) – \(\gamma\) is the coefficient for the fragmentation penalty used to promote spatial consistency in distinct open genomic regions and sparsity elsewhere. The ideal value of gamma is for a given dataset is dependent on the user’s preference. Increase to encourage solutions that are ‘block-like’.
beta (float) – Exponent for the denominator in the fragmentation penalty. If None, defaults to 1/2 when scale_gamma is True.
denom_const (float) – Constant to add to the denominator in the fragmentation penalty. If None, defaults to 0.5 when scale_gamma is True.
scale_gamma (bool) – If True, Scale the fragmentation penalty (γ) positionally based on the difference between adjacent scores. This will yield a more nuanced fragmentation penalty that does not discourage differences in adjacent decision variables if their corresponding scores are dissimilar and thus reflect a true change in state that should affect the solution: γ/(|Score_i - Score_i+1| + ε)^β See beta and denom_const for more details.
glop_parameters (parameters_pb2.GlopParameters) – glop-specific protocol buffer. If this is not None, the explicit solver arguments in this function definition are ignored. See https://protobuf.dev for more information on protocol buffers and parameters.proto for Google’s glop-specific protocol buffer.
glop_dual_feasibility_tolerance (float) – Dual feasibility tolerance for glop.
glop_primal_feasibility_tolerance (float) – Primal feasibility tolerance for glop.
glop_use_scaling (bool) – Use scaling for glop. If True, the scaling method is set to EQUILIBRATION and the cost scaling is set to CONTAIN_ONE_COST_SCALING. Recommended for most cases unless the user is confident the problem is not poorly-scaled. Future releases of ROCCO should support additional options.
glop_initial_basis (str) – Determines which of the glop-supported heuristics to identify an initial basis is executed. Options are ‘NONE’, ‘TRIANGULAR’, and ‘MAROS’. ‘TRIANGULAR’ is the default and recommended option.
threads (int) – Number of threads to use for optimization. Default is 0 (use default number of threads defined in solvers.proto). If threads is negative, the number of threads used will be the maximum of the number of available CPUs minus 1, or 1 in the case of a single-core machine.
verbose (bool) – Enable verbose output
save_model (str) – Save the model as an MPS file
glop_push_to_vertex (bool)
glop_allow_simplex_algorithm_change (bool)
- Returns:
Solution vector and optimal value
- Return type:
Tuple[np.ndarray, float]
- solve_relaxation_chrom_pdlp(scores, budget: float = 0.035, gamma: float = 1.0, beta: float | None = None, denom_const: float | None = None, pdlp_proto=None, pdlp_presolve_options_use_glop: bool = True, pdlp_termination_criteria_eps_optimal_absolute: float = 1e-08, pdlp_termination_criteria_eps_optimal_relative: float = 1e-08, pdlp_use_low_precision: bool = False, scale_gamma: bool = False, hint=None, threads: int = 0, verbose: bool = False, save_model: str | None = None) Tuple[numpy.ndarray, float][source]¶
Solve the relaxation for a specific chromosome using the first-order PDHG method, PDLP
See the full paper for PDLP (Applegate et al., 2021)
- Parameters:
scores (np.ndarray) – Scores for each genomic position within a given chromosome
budget (float) – \(b\) upper bounds the proportion of the chromosome that can be selected as open/accessible.
gamma (float) – \(\gamma\) is the coefficient for the fragmentation penalty used to promote spatial consistency in distinct open genomic regions and sparsity elsewhere. The ideal value of gamma is for a given dataset is dependent on the user’s preference. Increase to encourage solutions that are ‘block-like’.
beta (float) – Exponent for the denominator in the fragmentation penalty. If None, defaults to 1/2 when scale_gamma is True.
denom_const (float) – Constant to add to the denominator in the fragmentation penalty. If None, defaults to 1.0 when scale_gamma is True.
scale_gamma (bool) – If True, Scale the fragmentation penalty (γ) positionally based on the difference between adjacent scores. This will yield a more nuanced fragmentation penalty that does not discourage differences in adjacent decision variables if their corresponding scores are dissimilar and thus reflect a true change in state that should affect the solution: γ/(|Score_i - Score_i+1| + ε)^β See beta and denom_const for more details.
pdlp_proto (solvers_pb2.PrimalDualHybridGradientParams) –
pdlp-specific protocol buffer. If this is not None, the explicit solver arguments in this function definition are ignored. See https://protobuf.dev for more information on protocol buffers and solvers.proto for Google’s pdlp-specific protocol buffer.
pdlp_presolve_options_use_glop (bool) – Use glop’s presolve routines but solve with pdlp. Recommended for most cases unless computational resources are limited or the user has specified the problem already to exploit context-specific structure.
pdlp_termination_criteria_eps_optimal_absolute (float) – Appealing to strong duality for LPs, the duality gap (difference between the primal objective function and the dual objective function at a given iteration) must be less than this value plus a scaling of the primal/dual objective values (See solvers.proto linked above for exact details). If computational resources are limited, consider using 1.0e-4 per the ortools documentation.
pdlp_termination_criteria_eps_optimal_relative (float) –
Relative termination criterion for the pdlp solver. If computational resources are limited, consider using 1.0e-4 per the ortools documentation.
pdlp_use_low_precision (bool) – Use “loose solve”/low precision mode. This will override the termination arguments to weakened criteria (1.0e-4).
threads (int) – Number of threads to use for optimization. Default is 0 (use default number of threads defined in solvers.proto). If threads is negative, the number of threads used will be the maximum of the number of available CPUs minus 1, or 1 in the case of a single-core machine.
verbose (bool) – Enable verbose output
save_model (str) – Save the model as an MPS file
- Returns:
Solution vector and optimal value
- Return type:
Tuple[np.ndarray, float]
ROCCO: Readtracks¶
Basic suite of functions for calculating read density tracks from BAM files and collating data for downstream analysis.
- apply_filter(intervals: numpy.ndarray, count_matrix: numpy.ndarray, filter_type: str | None = None, savgol_window_bp: int | None = None, savgol_window_steps: int = 5, savgol_order: int | None = None, medfilt_kernel_bp: int | None = None, medfilt_kernel_steps: int = 5) Tuple[numpy.ndarray, numpy.ndarray][source]¶
Apply filtering to the count matrix.
- Parameters:
intervals (np.ndarray) – The genomic intervals.
count_matrix (np.ndarray) – The count matrix.
filter_type (str) – The type of filter to apply. Options are ‘savitzky-golay’ or ‘median’.
savgol_window_bp (int) – Window size for the Savitzky-Golay filter in base pairs.
savgol_window_steps (int) – Window size for the Savitzky-Golay filter in steps.
savgol_order (int) – Polynomial order for the Savitzky-Golay filter.
medfilt_kernel_bp (int) – Kernel size for the median filter in base pairs.
medfilt_kernel_steps (int) – Kernel size for the median filter in steps.
- Return type:
Tuple[numpy.ndarray, numpy.ndarray]
- apply_transformation(intervals: numpy.ndarray, count_matrix: numpy.ndarray, transform_log_pc: bool = False, log_const: float | None = None, transform_local_ratio: bool = False, local_ratio_window_bp: int | None = None, local_ratio_window_steps: int | None = None, local_ratio_pc: float | None = None, nonnegative: bool = True) Tuple[numpy.ndarray, numpy.ndarray][source]¶
Transform the count matrix after computing read densities.
- Parameters:
intervals (np.ndarray) – The genomic intervals.
count_matrix (np.ndarray) – The count matrix.
transform_log_pc (bool) – If True, apply a log2(x + c) transformation to data.
log_const (float) – Deprecated. Has no effect.
transform_local_ratio (bool) – If True, apply local ratio transformation to the values: x/(median_filter(x,row,kernel=local_ratio_window_bp) + ε).
local_ratio_window_bp (int) – Window size for the local ratio transformation in base pairs.
local_ratio_window_steps (int) – Window size for the local ratio transformation in steps.
local_ratio_pc (float) – Constant to add to the local reference before taking the ratio.
nonnegative (bool) – If True, the transformation will compute enrichment with respect to a non-negative reference. Otherwise, the transformation will allow negative reference values to divide the signal values.
- Returns:
A tuple containing two numpy arrays. The first array contains genomic intervals, and the second is a matrix (2d array) containing the corresponding values.
- Return type:
tuple(numpy.ndarray, numpy.ndarray)
- decompress_features_vals(intervals: numpy.ndarray, vals: numpy.ndarray, step: int)[source]¶
Reformat data from BedGraph-like intervals to have fixed-width, contiguous intervals. To avoid redundancy, methods generating BedGraph-like ouput will often create a single entry for repeated values over multiple adjacent intervals. For the use-cases in scope of this module, the savings achieved by this compressed representation are of little practical value and can be annoying to work with downstream. So this function decompresses the intervals and values.
- Parameters:
intervals (np.ndarray) – list-like of integer values corresponding to genomic positions
vals (np.ndarray) – list-like of floats representing the measured signal value at each interval
step (int) – integer specifying the step size in base pairs. ‘step’ is used synonymously with ‘bin size’.
- Returns:
A tuple of np.ndarray arrays: the first is the gap-filled intervals, the second is the signal values over the new intervals
- Return type:
tuple
- Seealso:
- Seealso:
- generate_bigwig(bam_file: str, step: int = 50, effective_genome_size: float = -1, norm_method: str = 'RPGC', min_mapping_score: int = -1, flag_include: int | None = None, flag_exclude: int = 3844, extend_reads: int = -1, center_reads: bool = False, ignore_for_norm: list | None = None, scale_factor: float = 1.0, ouput_file: str | None = None, output_file: str | None = None, num_processors: int = -1, overwrite: bool = False) str[source]¶
Generate a BigWig file from a BAM file.
- Parameters:
bam_file (str) – Path to the BAM file.
step (int) – Step size for the intervals. Used synonymously with ‘bin size’ throughout this documentation.
effective_genome_size (float) – Effective genome size for normalization. Required for RPGC normalization.
norm_method (str) – Normalization method. Must be one of: RPGC, RPKM, CPM, BPM. See deeptools documentation for more information.
min_mapping_score (int) – Minimum mapping score for reads to be included in the count track.
flag_include (int) – SAM flag to include in the count track. Equivalent to -f in samtools view.
flag_exclude (int) – SAM flag to exclude from the count track. Equivalent to -F in samtools view.
extend_reads (int) – Extend reads by a fixed number of base pairs. If < 0, ignore. If 0, extend reads to ‘estimated fragment length’. If > 0, extend reads by that number of base pairs.
center_reads (bool) – Center reads at the midpoint of the fragment length.
ignore_for_norm (list) – List of chromosome names to ignore for normalization.
scale_factor (float) – Factor to scale the values by.
output_file (str) – Path to the output BigWig file. If None, the file name will be generated based on the parameters used.
num_processors (int) – Number of processes to use for the calculation. If < 0, use all but one of the available processors.
overwrite (bool) – If True, overwrite the output file if it already exists. If False, skip the calculation if the output file already exists.
ouput_file (str | None)
- Returns:
Path to the output BigWig file.
- Return type:
str
- Raises:
FileNotFoundError – If the BAM file is not found.
ValueError – If the normalization method is not one of: RPGC, RPKM, CPM, BPM.
ValueError – If effective genome size is not specified and the normalization method is RPGC.
- generate_bigwigs(bam_files: list, step: int = 50, effective_genome_size: float = -1, norm_method: str = 'RPGC', min_mapping_score: int = -1, flag_include: int | None = None, flag_exclude: int = 3844, extend_reads: int = -1, center_reads: bool = False, ignore_for_norm: list | None = None, scale_factor: float = 1.0, num_processors: int = -1, overwrite: bool = True) list[source]¶
Generate BigWig files from a list of BAM files. This function is just a wrapper for
generate_bigwig()- Parameters:
bam_files (list) – List of paths to the BAM files.
step (int) – Step size for the intervals.
effective_genome_size (float) – Effective genome size for normalization. Required for RPGC normalization.
norm_method (str) – Normalization method. Must be one of: RPGC, RPKM, CPM, BPM. See deeptools documentation for more information.
min_mapping_score (int) – Minimum mapping score for reads to be included in the count track.
flag_include (int) – SAM flag to include in the count track. Equivalent to -f in samtools view.
flag_exclude (int) – SAM flag to exclude from the count track. Equivalent to -F in samtools view.
extend_reads (int) – Extend reads by a fixed number of base pairs. If < 0, ignore. If 0, extend reads to ‘estimated fragment length’. If > 0, extend reads by that number of base pairs.
center_reads (bool) – Center reads at the midpoint of the fragment length.
ignore_for_norm (list | None) – List of chromosome names to ignore for normalization.
scale_factor (float) – Factor to scale the values by.
num_processors (int) – Number of processes to use for the calculation. If < 0, use all but one of the available processors.
overwrite (bool) – If True, overwrite the output file if it already exists. If False, skip the calculation if the output file already exists.
- Returns:
List of paths to the output BigWig files.
- Return type:
list
- Raises:
FileNotFoundError – If a BAM file is not found.
ValueError – If the normalization method is not one of: RPGC, RPKM, CPM, BPM.
ValueError – If effective genome size is not specified and the normalization method is RPGC.
- Seealso:
- generate_chrom_matrix(chromosome: str, bigwig_files: list, chrom_sizes_file: str, step: int, const_scale: float = 1.0, round_digits: int = 5, scale_by_step: bool = False, filter_type: str | None = None, savgol_window_bp: int | None = None, savgol_window_steps: int = 5, savgol_order: int | None = None, medfilt_kernel_bp: int | None = None, medfilt_kernel_steps: int = 5, transform_log_pc: bool = False, log_const: float | None = None, transform_local_ratio: bool = False, local_ratio_window_bp: int | None = None, local_ratio_window_steps: int | None = None, local_ratio_pc: float | None = None)[source]¶
Create a matrix of read counts for a given chromosome from a list of BigWig files with potentially varying start and end positions. Provides options for transforming and filtering the tracks comprising the matrix.
- Parameters:
chromosome (str) – The chromosome to generate the matrix for.
bigwig_files (list) – List of paths to the BigWig files.
chrom_sizes_file (str) – Path to the chromosome sizes file.
step (int) – Step size for the intervals.
const_scale (float) – Factor to scale the signal values by after bigwig generation.
round_digits (int) – Number of decimal places to round the wig values to (not intervals).
scale_by_step (bool) – If True, scale the values by the step size.
filter_type (str) – Type of filter to apply. Options are ‘savitzky-golay’ or ‘median’.
savgol_window_bp (int) – Window size for the Savitzky-Golay filter in base pairs.
savgol_window_steps (int) – Window size for the Savitzky-Golay filter in steps.
savgol_order (int) – Polynomial order for the Savitzky-Golay filter.
medfilt_kernel_bp (int) – Kernel size for the median filter in base pairs.
medfilt_kernel_steps (int) – Kernel size for the median filter in steps.
transform_log_pc (bool) – If True, apply a log(x + 1) transformation to data.
log_const (float) – Deprecated. Has no effect.
transform_local_ratio (bool) – If True, apply local ratio transformation to the values. See
apply_transformation().local_ratio_window_bp (int) – Window size for the local ratio transformation in base pairs.
local_ratio_window_steps (int) – Window size for the local ratio transformation in steps.
local_ratio_pc (float) – Constant to add to the local reference before taking the ratio.
- Returns:
A tuple containing two numpy arrays. The first array contains genomic intervals, and the second contains the corresponding values.
- Return type:
tuple(numpy.ndarray, numpy.ndarray)
- Raises:
FileNotFoundError – If a BigWig file or chromosome sizes file is not found.
- Seealso:
- Seealso:
- Seealso:
- get_chrom_reads(bigwig_file: str, chromosome: str, chrom_sizes_file: str, step: int, const_scale: float = 1.0, round_digits: int = 5, scale_by_step: bool = False)[source]¶
Extract, decompress, and scale signal values (e.g., read counts) from a BigWig file for a specific chromosome.
- Parameters:
bigwig_file (str) – Path to the BigWig file.
chromosome (str) – Chromosome name to extract data for.
chrom_sizes_file (str) – Path to the chromosome sizes file.
step (int) – Step size for the intervals.
const_scale (float) – Factor to scale the signal values by after bigwig generation. Note, this is not the same parameter as the scale factor used in generate_bigwig.
round_digits (int) – Number of decimal places to round the wig values to (not intervals).
scale_by_step (bool) – If True, scale the values by the step size.
- Returns:
A tuple containing two np arrays. One for the genomic intervals, one for the values.
- Return type:
tuple
- Raises:
FileNotFoundError – If the BigWig file or chromosome sizes file is not found.
ValueError – If no non-zero values are found in the BigWig file.
- Seealso:
- Seealso:
- get_chroms_and_sizes(chrom_sizes_file)[source]¶
Parse a chromosome sizes file and return a dictionary of chromosome names and sizes. :param chrom_sizes_file: Path to the chromosome sizes file. :type chrom_sizes_file: str
- Returns:
A dictionary of chromosome names and sizes.
- Return type:
dict
- Raises:
FileNotFoundError – If the chromosome sizes file is not found.
- get_shape(matrix: numpy.ndarray) Tuple[source]¶
Helper function to get the shape of a 1D/2D numpy array
- Parameters:
matrix (numpy.ndarray)
- Return type:
Tuple
ROCCO: Scores¶
(Experimental) Compute post hoc peak statistics given samples’ BAM files and ROCCO peak output.
- get_ecdf(bam_files, length: int, chrom_sizes_file: str, nsamples=500, sample_scaling_constants: list | None = None, seed: int | None = None, null_stat: collections.abc.Callable[[numpy.ndarray], numpy.float64] = <function _null_stat>, trim_proportion: float = 0.005)[source]¶
Approximate null distribution of null_stat for a specific peak length by sampling genomic regions matched in length.
This function uses bedtools random to sample regions of a given length. Future implementations should account for other genomic features (e.g., mappability, GC content, etc.) when sampling background regions.
- Parameters:
bam_files (list or str) – List of paths to the BAM files OR a single filepath to a text file containing a list of BAM files (one filepath per line).
length (int) – Defines length of regions that will be randomly sampled to estimate the background distribution
chrom_sizes_file (str) – Filepath to chromosome sizes file.
nsamples (int) – Number of random ‘background’ regions to sample.
sample_scaling_constants (list) – List of scaling constants (floats) for each sample (provided by score_peaks() in current implementation).
seed (int) – Random seed supplied to bedtools random
null_stat (collections.abc.Callable[[numpy.ndarray], numpy.float64])
trim_proportion (float)
- Returns:
ECDF object with an evaluate() method.
- Return type:
scipy.stats._distn_infrastructure.rv_frozen
- Seealso:
score_peaks(), multi_ecdf()
- get_read_length(bam_file: str, num_reads: int = 1000, min_mapping_quality: int = 10)[source]¶
Get the mapped read length from a BAM file’s first num_reads.
- Parameters:
bam_file (str) – Path to the BAM file.
num_reads (int) – Number of reads to sample for the approximation.
min_mapping_quality (int)
- Returns:
estimated mapped read length
- Return type:
int
- multi_ecdf(bam_files, lengths, chrom_sizes_file: str, nsamples_per_length, sample_scaling_constants=None, seed=None, proc: int | None = None, null_stat: collections.abc.Callable[[numpy.ndarray], float] = <function _null_stat>)[source]¶
Compute ECDFs in parallel for each unique peak length
See get_ecdf() for details.
- Parameters:
chrom_sizes_file (str)
proc (int | None)
null_stat (collections.abc.Callable[[numpy.ndarray], float])
- raw_count_matrix(bam_files: str, peak_file: str, output_file: str, bed_columns: int = 3, overwrite=True)[source]¶
Generate a ‘raw’ count matrix from BAM files and a ROCCO output peak file.
- Parameters:
bam_files (list or str) – List of paths to the BAM files OR a single filepath to a text file containing a list of BAM files (one filepath per line).
bam_list_file (str) – list of filepaths to BAM files or a single filepath to a textfile containing a list of BAM files (one per line, order will be preserved in the output matrix).
peak_file (str) – Path to the BED file containing peaks.
output_file (str) – name of file to write the count matrix in.
bed_columns (int)
- Returns:
Name/path of the output file if successful, otherwise None.
- Return type:
str
Note
- Output Header.
The header of the output peak-count files (TSV) will preserve the order that the BAM files were supplied in.
Note
- BED3 format.
Default expects three-column BED peak files. If the peak file contains additional columns, set bed_columns accordingly.
- score_peaks(bam_files, chrom_sizes_file: str | None = None, peak_file: str | None = None, count_matrix_file: str | None = None, effective_genome_size: float | None = None, skip_for_norm: list = ['chrX', 'chrY', 'chrM'], row_scale=1000, ucsc_base=250, threads: int | None = None, pc=1, ecdf_nsamples=500, output_file='scored_peaks.bed', seed: int | None = None, proc: int | None = None, null_stat: collections.abc.Callable[[numpy.ndarray], float] = <function _null_stat>)[source]¶
Compute peak scores based on arcsinh-transformed, 1x-normalized counts, scaled by length. p-values based on ECDFs of read counts in each peak length.
- Parameters:
bam_files (list or str) – List of paths to the BAM files OR a single filepath to a text file containing a list of BAM files (one filepath per line).
chrom_sizes_file (str) – Path to the chromosome sizes file.
peak_file (str) – Path to (BED) file containing peak regions.
uscc_base (int) – Base value for UCSC score column. Default is 250 such that no peaks are indiscernible.
count_matrix_file (str | None)
effective_genome_size (float | None)
skip_for_norm (list)
threads (int | None)
seed (int | None)
proc (int | None)
null_stat (collections.abc.Callable[[numpy.ndarray], float])
ROCCO: Constants¶
Default effective genome sizes, chromosome size files, chromosome-specific budget/gamma parameters for supported assemblies: hg38, hg19, mm10, mm39, and dm6.
Universally optimal parameters/default values cannot be established for all datasets. When possible, users may find improved results by tuning the parameters to their specific data. See existing assembly-specific files for default parameter files you may use as a template.
Use the –params CLI argument to specify a custom parameter file.
Variables¶
- GENOME_DICTdict
Dictionary of genome assemblies with effective genome size, chromosome sizes file, and parameters file. Keys are assembly names, values are dictionaries with keys ‘effective_genome_size’, ‘sizes_file’, and ‘params’. Default assemblies: hg38, hg19, mm10, mm39, and dm6.